|
Special Topic: The Frauenfelder Protein Energy Landscape Model of Neutron Scattering (PELM) |
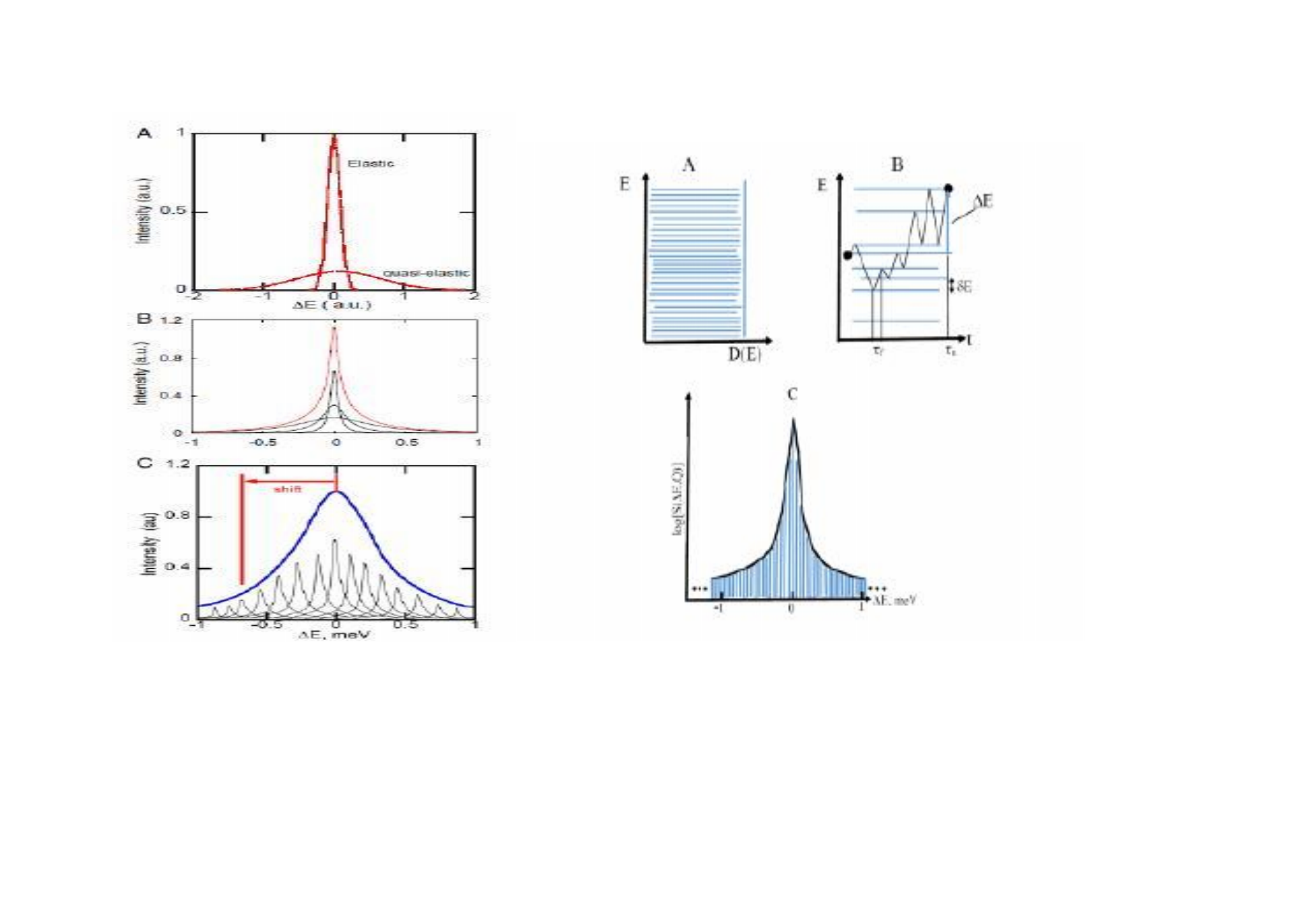
|
suggested reading: W. Doster, Are proteins dynamically heterogeneous?web
REPORTS
2023: HIRES2023 workshop: Resolution problems with biological samples. talk Doster
2022: QENS/WINS reports
1) Intracelluar molecular dynamics studied by neutron scattering by J. Zaccai et al.
EPJ Conf. Web 272 (2022): Comment Doster, Dosterebj08
2019: Franck-Condon picture of incoherent neutron scattering
by G. Kneller, PNAS 115, 94509455 (2018), attempt to justify energy landscape models of
proteins with quantum theoretical arguments.
Doster, PNAS Letter Apr. 2019,
G. Keller, Response to PNAS Letter Apr. 2019,
2018: Determination of dynamical heterogeneity of proteins from dynamic neutron scattering
Vural, Hong, Smith in Biophys. J. 114, 2397(2018)
Comment Doster 2017
Response by Vural to my review report and my back comments 2018 Comment Doster
related publication by Doster Are proteins dynamically heterogeneous?
2017: Low temperature decoupling of protein and water dynamics measured by neutron scattering
by A. Benedetto, Physical Chemistry Letters (2017)
Comment W. Doster, July 2019
2017: The role of momentum transfer during incoherent neutron scattering is explained
by the energy landscape model
by H. Frauenfelder, R.D. Young, P.W. Fenimore PNAS vol 114, 5130(2017).
Doster Comment:
the Frauenfelder zero Q elastic scattering effect reflects multiple scattering
and not the energy landscape
2015: Motional Displacement in Proteins, origin of wavevector-dependent values
by D. Vural, L. Hong, J. Smith, H. R. Glyde,
Phys.Rev. E. 91 052705(2015).
Comment Doster
2015 : Influence of Pressure and Crowding on the Subnanosecond Dynamics of Globular Proteins,
by M. Erlkamp, J. Marion, N. Martinez, C. Czeslik, J. Peters and R. Winter
J. Phys. Chem B 119 4842(2015).
Comment Doster
2014 : Wave mechanical model of incoherent quasielastic neutron scattering
in complex systems
by Hans Frauenfelder, Paul Fenimore and Robert Young,
PNAS , 111, 12764 (2014).
Wuttke: No case against scattering theory, PNAS Letter
Doster Comment
2014 : Does a dry protein undergo a glass transition by A. Frontzek, S. Strokov,
,J. Embs and S. Lushnikov, J. Phys. Chem B 118(11) 2791-2802(2014).
Comment
(The Freeze Drying Glass Transition in Dry Proteins)
2013 : Dynamics and Free Energy Landscape of Proteins, explored with the Mössbauer effect
and quasi-elastic neutron scattering by Frauenfelder, Young and Fenimore, J. Phys. Chem. 117 13301 (2013)
(The Mössbauer Model of Quasi-elastic Neutron Scattering)
Comment Doster
2013 : The Zaccai neutron resilence and site specific hydration dynamics in a globular protein by Miao, J. Smith et al.
Eur. J. Phys. E (2013) 36, 72.Comment Doster
2012/2013 : Change of caged dynamics of hydrated proteins by Capaccioli, Ngai, Ancherbak, Paciaroni,
J. Chem.Phys. 138 (2013) 235102.
Comment Doster
Two step scenario of the protein dynamical transition
2012: Puzzle of the Protein Dynamical Transition
by Magazu et al. J.Phys. Chem.B. (2011,2012)
Comment Doster
Evidence of coexistence of change of caged dynamics.. by Capaccioli, Ngai, Paciaroni,
J.Phys. Chem. B 116 (2012) 1745.
submitted open Comment by W. Doster, rejected by Editor of JPCB
2011 : The RENS puzzle
Elastic incoherent neutron scattering operating by varying
instrumental energy resolution: Principle, simulations,
and experiments of the resolution elastic neutron
scattering (RENS)
S. Magazu, F. Migliardo, A. Benedetto
Review of Scientific Instruments 82 (10), 105115 (2011)
nearly identical: Magazu, Migliardo, Benedetto, Vertessy in Chemical Physics 424(2013)26: Protein dynamics and neutron scattering..
Comment Doster
Comment Wuttke, Rev. Sci. Instr. 2011
2011 : Protein dynamical transition at 110 K,
by C. Kim, M. Tate and S. Gruner PNAS 108, 20897 (2011)
Comment
2011 : The Frauenfelder Mössbauer effect and the PDT:
Mössbauer effect in proteins, Young, Frauenfelder, Fenimore, PRL(2011)107, 158102
Comment Doster
2008 : A unified model of protein dynamics by Frauenfelder and Swenson, PNAS 2008
Comment Doster
2008 : Elliptical protein phase diagrams
Pressure and temperature dependent protein stability by Widersich, Skerra, Köhler, Friedrich,
PNAS 105, 575 (2008)
Comment Doster
2006 : Instrumental resolution effects interpreted as a fragile-strong crossover
Observation of fragile to strong dynamic cross-over of protein hydration water by
S.H. Chen, L.Liu, E. Fratini, P. Bagliaoni, A. Faraone and E. Mamontov,
PNAS USA 103, 9012 (2006)
Comment Doster
2004 : Frauenfelders alpha/beta relaxation
Bulk solvent and hydration shell fluctuations by Fenimore, Frauenfelder, Mc Mahon, Young
PNAS USA (2004)101,14408
Comment Doster
2003: Neutron Hydrogen Displacement Distribution in Myoglobin
Hydrogen atoms in proteins, Engler, Ostermann, Nijmura, Parak, PNAS USA (2003)100,10243
Comment Doster
2002 : Slaving II
Solvent fluctuations dominate protein dynamics and function
by Fenimore, Frauenfelder, Mc Mahon, Parak,
PNAS USA (2002)99,16047
Comment Doster
2002 : Confined water and the two simple explanation
A model for water motion in crystals of lysozyme based on an
incoherent quasi-elastic neutron scattering study by C.Bon, A.J. Dianoux, M. Ferrand and M.S. Lehmann,
Biophys. J. 83( 2002) 1578
Comment Doster
2002 : The protein dynamical transition may have a simple explanantion
by M. R. Daniel, J. Finney and J. Smith, Faraday Discussion (2002) 122,163
Comment Doster
2000: Protein force constants from elastic displacements?
How soft is a protein? A protein dynamics force constant measured by neutron scattering by J. Zaccai,
Science 288,1604( 2000)
Comment Doster
1998 : Dynamic labelling of different functional parts of BR
by V. Reat, H. Patzelt, M. Ferrand, C. Pfister, D. Oesterhelt, G. Zaccai PNAS 95(1998)4970
Comment Doster
1998 : Activity below the dynamic transition?
Enzyme Activity below the Protein Dynamical Transition at 220K by R. Daniel,
J.Smith, M. Ferrand, S. Hery, R. Dunn, J. Finney, Biophys. J. 75 (1998) 2504
Comment Doster
1993 : Melting of a frozen protein solution
Thermal motion and function of bacteriorhodopsin in purple membrane, effect of temperature and
hydration observed by neutron scattering by M. Ferrand, A. Dianoux, W. Petry an G. Zaccai,
PNAS 90, 9668 (1993)communicated by Hans Frauenfelder.
Comment Doster
1992 : Confined water (I):
Single particle dynamics of hydration water in protein, M.C. Bellissent-Funel,
J. Teixera, J.F. Bradley, S.H. Chen and L. Crespi, Physica B 181 &181, 740 (1992).
Comment
1991 : Review Article on MD Simulation and Experiments
Protein Dynamics: comparison of simulations with inelastic neutron
scattering experiments, by J. Smith, Quat. Rev. Biophys.24 (1991), 227
Comment Doster
1991 : Frauenfelders Energy Landscapes
The energy landscapes and motions in proteins, H. Frauenfelder, S. Sligar and P. Wolynes,
Science 254 (1991) 1598
Comment
1990 : Vacuum simulation of a hydrated protein
Dynamics of myoglobin: comparison of simulation results with neutron scattering spectra,
by J. Smith, K. Kuczera and M. Karplus, PNAS USA (1990)87, 1601.
Comment Doster
1990 : First simulation of a hydrated protein: The temperature dependence of dynamics of hydrated myoglobin, comparison of force field calculations
with neutron scattering data
by R. Loncharich and B. Brooks, J. Mol. Biol. (1990)215, 439
1989: first spectral analysis of protein dynamics:
Dynamical transition of myoglobin revealed by inelastic neutron scattering, W. Doster, W. Petry and
S. Cusack, Nature 337,754(1989)
Comment Doster
Internal dynamics of globular proteins, comparison of neutron scattering measurements and theoretical models
by J. Smith, K. Kuczera, B. Tidor, W. Doster, S.Cusack and M. Karplus, Physica B(1989) 156, 437.
Comment Doster
1982: Ligand Binding to Hexokinase
Inelastic neutron scattering analysis of hexokinase dynamics and its modification on binding of
glucose by B. Jacrot, S. Cusack, A. Dianoux and D. Engelman, Nature 300 (1982)84
Comment Doster
1980/1996 : spurious oscillations of hydration water jump rate
Molecular dynamics of hydrated proteins, H. Middendorf, J. Randall and A. J. Leadbetter,
Phil. Trans. R. Soc. Lond.B. (1980) 290, 639. and Middendorf, Phys. B. 226, 113 (1996)
Comment Doster